Entries in Liston lab (248)
Greater understanding of immune signalling molecule raises hope for improved clinical use
 Tuesday, June 14, 2022 at 5:20PM
Tuesday, June 14, 2022 at 5:20PM Source of immune signals alters the immune response
Key points:
- Researchers have identified source-specific effects of the signalling molecule interleukin 2 (IL2) on the immune response.
- IL2 is an important signalling molecule that has been harnessed as a biologic therapy for a number of diseases but can result in unwanted side-effects.
- This study, conducted using new mouse models, found that the immune response to IL2 is dependent on the cellular source of the IL2 production.
- Their new insight explains the link between IL2 treatments and side-effects, opening up the potential to apply this powerful immune modulator to optimise treatments while avoiding off-target effects.
A detailed update to our understanding of the key immune system signalling molecule interleukin 2 has been published today by researchers at the Babraham Institute. Their findings explain common side effects of IL2-based therapies, and identify potential new uses of IL2 as an immune-modulating biologic drug. This research was only possible thanks to a new mouse model which allowed researchers to control which immune cell types produced IL2. With further research, this understanding of the rules dictating which cells respond to IL2 could allow scientists to optimise autoimmune and cancer treatment while avoiding unwanted side-effects.
Dr Carly Whyte, lead author on the paper who undertook this research as a postdoctoral researcher in the Liston lab, said: "IL2 is a protein that is normally tightly regulated in the immune system because it has such strong effects. However, when IL2 is given as a therapeutic treatment, these normal restrictions on IL2 are overruled. By using mouse models, we have found that the presence of IL2 in certain zones of the immune system leads to some of the same side-effects that we see in human patients treated with IL2. We hope that by understanding more about how IL2 works in different zones, this treatment might be tailored to be more effective."
IL2 is involved in a large number of different communication networks in the immune system, being produced by a variety of cellular sources and affecting a diversity of cell ‘responders’. It is not only needed for maintaining regulatory T cells, which prevent our body’s immune system from attacking itself, but also CD8 T cells, which attack tumour cells and virus-infected cells. Owing to this dual functionality, IL2 has been harnessed to both promote an immune response, and limit one, depending on the target cells. Despite being actively explored in hundreds of ongoing clinical trials, the full therapeutic potential is currently limited by frequently-encountered side-effects.
Previous explanations for these side-effects were based on the high doses of IL2 when given as a biologic drug, but Prof. Adrian Liston and his team were able to demonstrate that the cell-type making IL2, and the location of those cells, dramatically change the consequences of IL2 exposure. Dr Kailash Singh, co-lead author, explains "Our genetically modified mouse models showed that the immune responses are varied depending on the source of IL2. Our findings revealed that the IL2 response is very much context-dependent, and is not solely due to the concentration of IL2."
Prof. Liston, a senior group leader in the Institute’s Immunology research programme, said: “This work changes the way we think about IL2 as a decades-old therapeutic molecule, demonstrating that it is not just the dose of the IL2 that matters, but also where it is located in the body. Putting together the pieces of this cause and effect intricacy has involved several remarkable scientists and over a decade of research. It was only by bringing together experts in animal research, flow cytometry and immunology, that we had the know-how to tackle the complexity of this question. We’re increasingly aware of the therapeutic power of the immune system, and these findings provide a new avenue of investigation for designing biologic drugs.”
Dr James Dooley, joint senior author of the study, said: "The next generation of biologics will be smarter and tailored to the biology of the disease. This work teaches us that one route of smart design of IL2 is to target delivery to different parts of the body, potentially allowing us to drive very different therapeutic outcomes in patients."
Read the original paper at The Journal of Experimental Medicine!
 Liston lab,
Liston lab,  immunology
immunology CYTO 2022 presentation by Dr Oliver Burton
 Friday, June 10, 2022 at 10:57AM
Friday, June 10, 2022 at 10:57AM  Liston lab,
Liston lab,  data science,
data science,  immunology
immunology Career milestone: 200 papers
 Saturday, May 28, 2022 at 10:00AM
Saturday, May 28, 2022 at 10:00AM Our new paper out at Nature Immunology was my 200th scientific paper! A good time to look back on the portfolio.
First, my coauthors:
 My most frequent coauthor is James Dooley, no surprise since we've run the lab together these last 14 years! 67 articles coauthored, a good third of my papers. We've had a lot of staff and students trained in our lab over these years (165, to be exact), including a few who changed the direction of our lab - Stephanie Humblet-Baron coauthored 46 papers and Susan Schlenner coauthored 17, both are now professors at the University of Leuven. Vaso Lagou also coauthored 17 papers as a post-doc, before moving over to the Sanger. Jossy Garcia-Perez and Dean Franckaert were both PhD students, with 15 coauthorships, now working together at CellCarta. Our major collaborators come through clearly: Carine Wouters (28 papers) and Isabelle Meyts (15 papers) on the clinical side, An Goris (15 papers) on genetics, and Michelle Linterman (18 papers), Patrick Matthys (16 papers) and Sylvie Lesage (11 papers) for immunology.
My most frequent coauthor is James Dooley, no surprise since we've run the lab together these last 14 years! 67 articles coauthored, a good third of my papers. We've had a lot of staff and students trained in our lab over these years (165, to be exact), including a few who changed the direction of our lab - Stephanie Humblet-Baron coauthored 46 papers and Susan Schlenner coauthored 17, both are now professors at the University of Leuven. Vaso Lagou also coauthored 17 papers as a post-doc, before moving over to the Sanger. Jossy Garcia-Perez and Dean Franckaert were both PhD students, with 15 coauthorships, now working together at CellCarta. Our major collaborators come through clearly: Carine Wouters (28 papers) and Isabelle Meyts (15 papers) on the clinical side, An Goris (15 papers) on genetics, and Michelle Linterman (18 papers), Patrick Matthys (16 papers) and Sylvie Lesage (11 papers) for immunology.

The research topics come out via the key words from the titles. Tregs, Foxp3, T cells and the thymus all leap out, but looking closely you'll see pretty much every branch of immunology represented. A special call-out to my favourite cytokine, IL2 (with 14 papers, and getting stronger) and our microRNA papers (22 papers, but it was just a phase).
The work is pretty evenly split between mouse and human, although we tend to use "mouse" a lot more in the title. In terms of topics, 88 papers work on autoimmune diseases, with 17 touching on diabetes. 40 papers intersect with cancer biology or cancer immunology, 30 papers are on primary immunodeficiencies (across both mouse and human, but spread out over so many genes and syndromes they don't pop out here). 15 papers papers are on neuroimmunology, a current strength of the lab.
 Finally, the journals that published our work! Really thrilled to see Nature Immunology up there, with 8 papers. We have a scattering of other top journals, Cell, Nature, Science, Nature Medicine, Nature Neuroscience, Nature Genetics all get a mention. Our most popular journals, however, are The Journal of Allergy and Clinical Immunology (10 papers), which has published some of our key work on primary immunodeficiency in both mouse and human, and Immunology and Cell Biology (10 papers), the Australian and New Zealand society journal (and a pleasure to work with!). I've been told by senior researchers that publishing anything below the top tier "dilutes my record", but I'm proud of all the science we do, and work to make sure that every story finds a home and every staff member or student with data gets to show it on the international stage.
Finally, the journals that published our work! Really thrilled to see Nature Immunology up there, with 8 papers. We have a scattering of other top journals, Cell, Nature, Science, Nature Medicine, Nature Neuroscience, Nature Genetics all get a mention. Our most popular journals, however, are The Journal of Allergy and Clinical Immunology (10 papers), which has published some of our key work on primary immunodeficiency in both mouse and human, and Immunology and Cell Biology (10 papers), the Australian and New Zealand society journal (and a pleasure to work with!). I've been told by senior researchers that publishing anything below the top tier "dilutes my record", but I'm proud of all the science we do, and work to make sure that every story finds a home and every staff member or student with data gets to show it on the international stage.
 Liston lab,
Liston lab,  science careers
science careers Harnessing the immune system to treat traumatic brain injury
 Thursday, May 26, 2022 at 5:51PM
Thursday, May 26, 2022 at 5:51PM Pioneering new treatment leads to improved recovery from brain trauma in mice
Research offers new approach to limiting harmful brain inflammation after injury or disease
- Researchers have designed a targeted therapeutic treatment that restricts brain inflammation. The effectiveness of this approach in improving outcomes was demonstrated following brain injury, stroke or multiple sclerosis in mice.
- The system increases the number of regulatory T cells, mediators of the immune system’s anti-inflammatory response, in the brain.
- By boosting the number of T regulatory cells in the brain, the researchers were able to prevent the death of brain tissue in mice following injury and the mice performed better in cognitive tests.
- The treatment has a high potential for use in patients with traumatic brain injury, with few alternatives currently available to prevent harmful neuroinflammation.
A therapeutic method for harnessing the body’s immune system to protect against brain damage is published today by researchers from the Babraham Institute’s Immunology research programme. The collaboration between Prof. Adrian Liston (Babraham Institute) and Prof. Matthew Holt (VIB and KU Leuven; i3S-University of Porto) has produced a targeted delivery system for boosting the numbers of specialised anti-inflammatory immune cells specifically within the brain to restrict brain inflammation and damage. Their brain-specific delivery system protected against brain cell death following brain injury, stroke and in a model of multiple sclerosis. The research is published today in the journal Nature Immunology.
Traumatic brain injury, like that caused during a car accident or a fall, is a significant cause of death worldwide and can cause long-lasting cognitive impairment and dementia in people who survive. A leading cause of this cognitive impairment is the inflammatory response to the injury, with swelling of the brain causing permanent damage. While inflammation in other parts of the body can be addressed therapeutically, but in the brain it problematic due to the presence of the blood-brain barrier, which prevents common anti-inflammatory molecules from getting to the site of trauma.
Prof. Liston, a senior group leader in the Institute’s Immunology programme, explained their approach: “Our bodies have their own anti-inflammatory response, regulatory T cells, which have the ability to sense inflammation and produce a cocktail of natural anti-inflammatories. Unfortunately there are very few of these regulatory T cells in the brain, so they are overwhelmed by the inflammation following an injury. We sought to design a new therapeutic to boost the population of regulatory T cells in the brain, so that they could manage inflammation and reduce the damage caused by traumatic injury.”
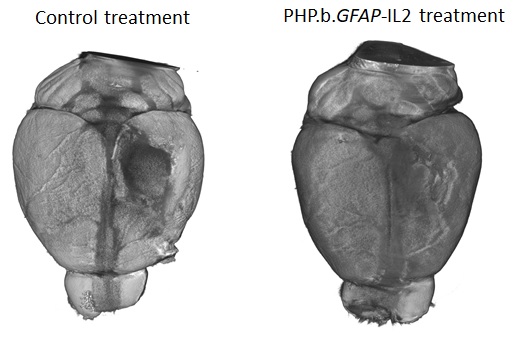
The research team found that regulatory T cell numbers were low in the brain because of a limited supply of the crucial survival molecule interleukin 2, also known as IL2. Levels of IL2 are low in the brain compared to the rest of the body as it can’t pass the blood-brain barrier.
Together the team devised a new therapeutic approach that allows more IL2 to be made by brain cells, thereby creating the conditions needed by regulatory T cells to survive. A ‘gene delivery’ system based on an engineered adeno-associated viral vector (AAV) was used: this system can actually cross an intact blood brain barrier and deliver the DNA needed for the brain to produce more IL2 production.
“Even though the number of Treg cells in the naive CNS is very low, they are a powerful immunosuppressive tool that we exploited to reduce damage-induced inflammation without harnessing the brain.” explains the first author, Dr. Pasciuto.
Commenting on the work, Prof. Holt said ‘For years, the blood-brain barrier has seemed like an insurmountable hurdle to the efficient delivery of biologics to the brain. Our work, using the latest in viral vector technology, proves that this is no longer the case; in fact, it is possible that under certain circumstances, the blood-brain barrier may actually prove to be therapeutically beneficial, serving to prevent ‘leak’ of therapeutics into the rest of the body’.
The new therapeutic designed by the research teams was able to boost the levels of the survival molecule IL2 in the brain, up to the same levels found in the blood. This allowed the number of regulatory T cells to build up in the brain, up to 10-fold higher than normal. To test the efficacy of the treatment in a mouse model that closely resembles traumatic brain injury accidents, mice were given carefully controlled brain impacts and then treated with the IL-2 gene delivery system. The scientists found that the treatment was effective at reducing the amount of brain damage following the injury, assessed by comparing both the loss of brain tissue and the ability of the mice to perform in cognitive tests.
Lead author, Dr Lidia Yshii, explained: “Seeing the brains of the mice after the first experiment was a ‘eureka moment’ – we could immediately see that the treatment reduced the size of the injury lesion”.
Recognising the wider potential of a drug capable of controlling brain inflammation, the researchers also tested the effectiveness of the approach in experimental mouse models of multiple sclerosis and stroke. In the model of multiple sclerosis, treating mice during the early symptoms prevented severe paralysis and allowed the mice to recover faster. In a model of stroke, mice treated with the IL2 gene delivery system after a primary stroke were partially protected from secondary strokes occurring two weeks later. In a follow-up study, still undergoing peer review, the research team also demonstrated that the treatment was effective at preventing cognitive decline in ageing mice.
“By understanding and manipulating the immune response in the brain, we were able to develop a gene delivery system for IL-2 as a potential treatment for neuroinflammation. With tens of millions of people affected every year, and few treatment options, this has real potential to help people in need. We hope that this system will soon enter clinical trials, essential to test whether the treatment also works in patients." said Prof. Liston.
Dr Ed Needham, a neurocritical care Consultant at Addenbrooke’s Hospital who was not a part of the study, commented on the clinical relevance of these results: “There is an urgent clinical need to develop treatments which can prevent secondary injury that occurs after a traumatic brain injury. Importantly these treatments have to be safe for use in critically unwell patients who are at high risk of life-threatening infections. Current anti-inflammatory drugs act on the whole immune system, and may therefore increase patients' susceptibility to such infections. The exciting progress in this study is that, not only can the treatment successfully reduce the brain damage caused by inflammation, but it can do so without affecting the rest of the body's immune system, thereby preserving the natural defences needed to survive critical illness."
 Liston lab,
Liston lab,  Medicine,
Medicine,  immunology,
immunology,  neuroscience
neuroscience Baanbrekende nieuwe behandeling leidt tot beter herstel na hersentrauma bij muizen
 Thursday, May 26, 2022 at 5:00PM
Thursday, May 26, 2022 at 5:00PM Onderzoekers hebben een gerichte therapie ontworpen die ontsteking in de hersenen tegengaat. De aanpak – waarbij doelgericht DNA tot in de hersenen wordt gebracht - blijkt succesvol bij muizen met een hersenletsel, beroerte of multiple sclerose
Kort & bondig:
- Onderzoekers hebben een gerichte therapie ontworpen die ontsteking in de hersenen tegengaat. De aanpak – waarbij doelgericht DNA tot in de hersenen wordt gebracht - blijkt succesvol bij muizen met een hersenletsel, beroerte of multiple sclerose.
- De behandeling bestaat er in het aantal regulatoire T-cellen, die de anti-inflammatoire respons van het immuunsysteem reguleren, te verhogen in de hersenen.
- Door het aantal regulerende T-cellen in de hersenen te verhogen, konden de onderzoekers het afsterven van hersenweefsel bij muizen na verwonding voorkomen en deden de muizen het beter bij cognitieve testen.
- Voor patiënten met een traumatisch hersenletsel zijn er momenteel weinig opties om schadelijke neuroinflammatie te voorkomen. De nieuwe resultaten zijn dus erg hoopgevend.
Onderzoekers uit Leuven en uit het Verenigd Koninkrijk publiceren vandaag nieuwe resultaten over een therapeutische piste waarbij het immuunsysteem wordt ingezet om hersenbeschadiging te voorkomen. De samenwerking tussen professor Adrian Liston (voorheen VIB en KU Leuven en sinds enkele jaren verbonden aan het Babraham Institute in het VK) en professor Matthew Holt (verbonden aan VIB, KU Leuven en de i3S-Universiteit van Porto) leidde tot een systeem om in de hersenen het aantal gespecialiseerde ontstekingsremmende immuuncellen op te voeren om op die manier hersenontsteking en -beschadiging te beperken. De aanpak bleek alvast in muismodellen tot minder hersenschade te leiden na hersenletsel, beroerte of bij multiple sclerose. Het onderzoek wordt vandaag gepubliceerd in het tijdschrift Nature Immunology.
Traumatisch hersenletsel, zoals veroorzaakt na een auto-ongeval of een val, is wereldwijd een belangrijke doodsoorzaak. Het kan langdurige en ernstige gevolgen hebben voor mensen die het overleven, onder de vorm van cognitieve problemen en zelfs dementie. Een belangrijke oorzaak van deze cognitieve stoornissen is de ontstekingsreactie op het letsel. Hierbij veroorzaakt zwelling van de hersenen permanente schade. Terwijl ontsteking in andere delen van het lichaam kan worden aangepakt met geneesmiddelen, is dit in de hersenen heel moeilijk door de aanwezigheid van de bloed-hersenbarrière, die voorkomt dat gewone ontstekingsremmende moleculen op de plaats van het (hersen)trauma kunnen komen.
Prof. Liston: "Ons lichaam heeft zijn eigen ontstekingsremmende respons: regulatoire T-cellen zijn in staat om ontstekingen waar te nemen en een cocktail van natuurlijke ontstekingsremmers te produceren. Helaas zijn er maar heel weinig van deze regulerende T-cellen in de hersenen. Wij probeerden een nieuw therapeutisch middel te ontwikkelen om de hoeveelheid regulerende T-cellen in de hersenen te vergroten. Indien er voldoende regulatoire T-cellen zijn, zo redeneerden we, zouden ze de ontsteking na een letsel kunnen beheersen en de schade beperken."
Het onderzoeksteam ontdekte dat het aantal regulerende T-cellen in de hersenen laag was door een beperkte aanvoer van het cruciale overlevingsmolecuul interleukine 2, ook bekend als IL2. Het niveau van IL2 is laag in de hersenen vergeleken met de rest van het lichaam omdat het niet doorheen de bloed-hersenbarrière kan.
Samen bedacht het team een nieuwe therapeutische aanpak waardoor meer IL2 kan worden aangemaakt door hersencellen. De onderzoekers gebruikten een "gene delivery"-systeem op basis van een virale vector: dit systeem kan daadwerkelijk een intacte bloed-hersenbarrière passeren en het DNA afleveren dat de hersenen nodig hebben om meer IL2 aan te maken.
Prof. Holt: "Jarenlang leek de bloed-hersenbarrière een onoverkomelijke hindernis voor de efficiënte toediening van biologische geneesmiddelen in de hersenen. Ons werk, waarbij we gebruik maken van de nieuwste virale vectortechnologie, bewijst dat dit niet langer het geval is; het is zelfs mogelijk dat de bloed-hersenbarrière onder bepaalde omstandigheden therapeutisch gunstig kan zijn, omdat ze – eenmaal op hun bestemming – het 'lekken' van geneesmiddelen naar de rest van het lichaam verhindert."
Dankzij hun aanpak waren de onderzoekers in staat de niveaus van de overlevingsmolecule IL2 in de hersenen op te voeren tot dezelfde niveaus als in het bloed. Hierdoor kon het aantal regulerende T-cellen zich in de hersenen opbouwen, tot 10 maal hoger dan normaal. Om de doeltreffendheid van de behandeling te testen in muizen. Wat bleek? Muizen met meer IL2 hadden inderdaad minder hersenschade na een letsel en presteerden ook beter in cognitieve tests.
Dr. Lidia Yshii, van het team aan KU Leuven, legt uit: "Toen we de hersenen van de muizen zagen na het eerste experiment, was dit een echt 'eureka-moment' - we zagen meteen dat de behandeling het letsel had verkleind."
De onderzoekers testten ook de doeltreffendheid van hun aanpak in experimentele muismodellen voor multiple sclerose en beroerte—met succes. In een vervolgstudie, die nog aan peer review wordt onderworpen en dus nog niet gepubliceerd is, toont het onderzoeksteam ook aan dat de behandeling doeltreffend was om cognitieve achteruitgang bij ouder wordende muizen te voorkomen.
"Door de immuunrespons in de hersenen te begrijpen en erop in te spelen, waren we in staat een gen-toedieningssysteem voor IL-2 te ontwikkelen als een potentiële behandeling voor neuro-inflammatie. Met tientallen miljoenen mensen die er elk jaar mee te maken krijgen en met bovendien weinig beschikbare behandelingsmogelijkheden, biedt onze nieuwe aanpak reële mogelijkheden om mensen in nood te helpen. We hopen dat dit systeem binnenkort aan klinische proeven zal worden onderworpen, die essentieel zullen zijn om te testen of de behandeling ook bij patiënten werkt," aldus Prof. Liston.
Dr. Ed Needham, een neuroloog in het Addenbrooke's Hospital in het VK die geen deel uitmaakte van de studie, gaf commentaar op de klinische relevantie van deze resultaten: "Er is een dringende klinische noodzaak om behandelingen te ontwikkelen die secundair letsel kunnen voorkomen dat optreedt na een traumatisch hersenletsel. Belangrijk is dat deze behandelingen veilig zijn voor gebruik bij kritisch zieke patiënten die een hoog risico lopen op levensbedreigende infecties. De huidige ontstekingsremmende geneesmiddelen werken in op het gehele immuunsysteem en kunnen daardoor de gevoeligheid van patiënten voor dergelijke infecties vergroten. Wat deze studie zo interessant maakt is dat de behandeling niet alleen met succes de door ontsteking veroorzaakte hersenschade kan verminderen, maar dat zij dit kan doen zonder de rest van het immuunsysteem van het lichaam aan te tasten, waardoor de natuurlijke afweer die nodig is om kritieke ziekte te overleven, behouden blijft."
 Liston lab,
Liston lab,  Medicine,
Medicine,  immunology,
immunology,  neuroscience
neuroscience The genetic underpinnings of severe staph infections
 Friday, May 20, 2022 at 10:09AM
Friday, May 20, 2022 at 10:09AM In a large collaborative effort, an international team of researchers describes a genetic mutation that predisposes individuals to severe staphylococcus infections. The research, in collaboration with the Babraham Institute, appears in the latest edition of Science.
Staphylococcus aureus is usually harmless. Many of us host colonies of this bacterium in our noses and on our skin without suffering more than the occasional rash. But some strains of staph—particularly MRSA—can turn deadly, leading to pneumonia and sepsis that claims 20,000 lives in the U.S. each year. Now a new study describes a mutation that predisposes some individuals to severe staph infection.
Babraham Institute researcher Adrian Liston collaborated with Belgian doctors Isabelle Meyts, Rik Schrijvers, and Carine Wouters, to investigate the genetic and immunological cause of the disease.
The research, published in Science, describes a mutation in the OTULIN gene in patients who suffer life-threatening staph infections. “In a collaborative effort, several unrelated patients were identified with identical severe clinical presentations and we found a common genetic cause of the disease”, says Frederik Staels, MD, and PhD student at the University of Leuven in Belgium.
"We have characterized severe Staphylococcus aureus infection at the genetic, cellular, immunological, and clinical levels," says Dr. András Spaan, a clinical microbiologist working at The Rockefeller University in New York, who was one of the coordinators of this large international effort. "By integrating these levels, we have been able to establish causality and provide clues for future pharmaceutical interventions."
The study indicated that about 30 percent of people with this OTULIN mutation develop severe disease. This risk was reduced in patients that had acquired specific anti-staph antibodies, while patients without such neutralizing antibodies remained at very high risk of developing severe infections. “This is a potential path to protecting this at-risk patients”, explains Prof Adrian Liston, “the protected patients had acquired anti-staph antibodies through natural exposure, with each exposure being a high-risk gamble for life-threatening infection. If these patients can be identified and vaccinated, the anti-staph antibodies they gain through vaccination may protect them from serious illness”.
"Studies on these disorders can act as a compass," Prof Humblet-Baron, University of Leuven, says, “They bring new mechanistic insights about the interaction between hosts and pathogens, which can also benefit the general population with better understanding about staph infection pathogenesis.”
“From a clinical point of view, this work is of great relevance to physicians confronted with patients manifesting severe, life-threatening episodes of skin and/or lung inflammation, necessitating prompt recognition and treatment,” says Prof. Wouters, pediatric rheumatologist at the University of Leuven.
Read the paper over at Science.
 Liston lab,
Liston lab,  Medicine,
Medicine,  immunology
immunology A Cross Entropy test for tSNE by Dr Oliver Burton
 Tuesday, May 17, 2022 at 3:07PM
Tuesday, May 17, 2022 at 3:07PM The low-dimensional representation of high-dimensional data makes t-SNE an attractive visualisation tool, yet it also has value as an analytical tool. We have developed the Cross Entropy test, a statistical test capable of distinguishing biological differences in single cell t-SNE representations, while being robust against false detection of differences in technical replicates or the seed-dependent variation in t-SNE generation. As the t-SNE algorithm is driven by the cross entropy of the individual cells in the dataset, and the t-SNE fixes the average point entropy, each t-SNE can be considered a distribution of cross entropy divergences. Deriving a distribution of cross entropy divergences per t-SNE plot therefore allows the use of the Kolmogorov-Smirnov test to evaluate the degree of difference between two, or more, t-SNE plots.
The Cross Entropy test is a useful tool for calculating p values on the difference between any two t-SNE or UMAP plots, whether the data comes from flow cytometry, mass cytometry or single cell sequencing. Further, the test generates a quantitative comparison of the extent of differences, allowing you to compare multiple t-SNE or UMAP plots and identify outgroups and clustered samples. See an overview of the Cross Entropy test given by Dr Oliver Burton:
 Liston lab,
Liston lab,  data science
data science Our paper discussed on RheumMadness podcast
 Sunday, March 6, 2022 at 3:51PM
Sunday, March 6, 2022 at 3:51PM The RheumMadness podcast scouted our paper on using machine learning and immunoprofiling to understand juvenile idiopathic arthritis. Their summary?
Future: Future implications for immunophenotyping machine learning include the diagnosis, treatment and prognosis of all rheumatologic conditions. With the increase in potential immunomodulating targeted therapies, along with the classification of disease based on those same immune targets, an exciting possibility of choosing precise individualized treatment plans for our patients exists.In pediatric rheumatology, we are accustomed to using complicated clinical algorithms to properly diagnose and treat our patients. But is this really the most accurate system? Machine learning and immunophenotyping have the potential to turn the field inside out.Chances in the Tournament: As the only pediatric team in play, this team is the dark horse. However, the long-term clinical implications of this team are arguably more far-reaching than any other team in the Machine Region—and the entire tournament. Despite the small number of participants in this study, the exclusion of psoriatic and enthesitis-related JIA, and the lack of attention given to race, ethnicity and environmental factors that could potentially alter immune signatures, we still believe the strengths of this article make it a crucial one. We stand an excellent chance.Immunophenotyping machine learning has implications for more than just anti-tumor necrosis factor response in RA, like our opponents would argue. Our study shows that its implications stretch far beyond one diagnosis or two therapy choices. In fact, pediatric rheumatologists have just begun to pave the way for better classifying patients in the adult world as well. Could eight subtypes of RA actually exist, and you just don’t know it yet? Immunophenotyping through machine learning could be the disrupter you’ve been waiting for. This study can go all the way.
 Liston lab,
Liston lab,  immunology
immunology 





