AutoSpill: a method for calculating spillover coefficients in high-parameter flow cytometry
 Wednesday, July 1, 2020 at 11:06AM
Wednesday, July 1, 2020 at 11:06AM I am really thrilled to release AutoSpill onto BioRxiv. AutoSpill is a novel method for applying compensation to flow cytometry data, which reduces the error by ~100,000-fold. It is thanks to AutoSpill that we can push machines to their max colours, and actually get good quality 40+ parameter flow cytometry data. AutoSpill is a beautiful example of what maths can add to #immunology, led by the talented Dr Carlos Roca.

So how does AutoSpill work? If you just want to compensate your data, simply upload your single colour controls to https://autospill.vib.be and then copy the spillover matrix to your flow cytometry program of choice. Dr Carly Whyte made this easy two minute tutorial:
If you program your flow cytometry analysis in R, we have also released the AutoSpill full code, so you can add this to your bioinformatics pipeline.
Here are a few examples of the error reduction you can get with AutoSpill:
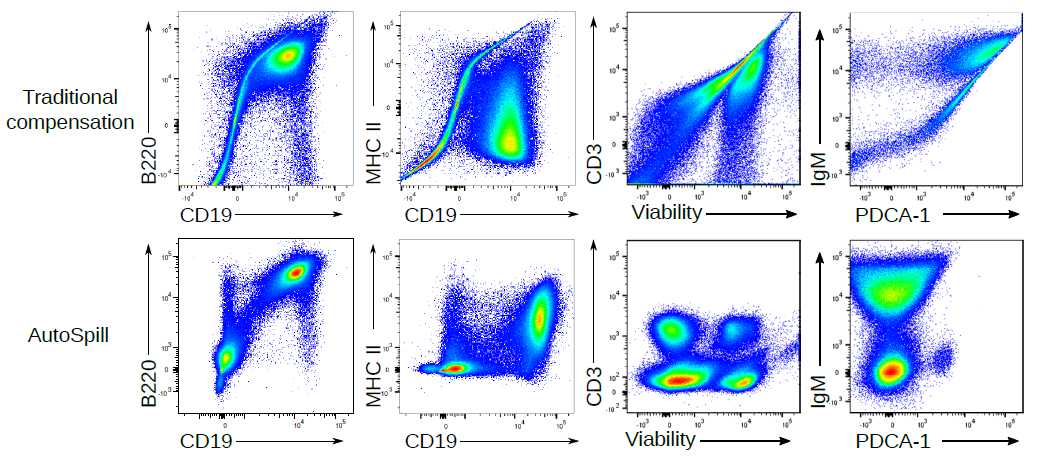
In high dimensional flow cytometry traditional compensation errors create artefacts. AutoSpill creates a perfect spillover matrix. What does a "perfect" spillover matrix mean? An error reduction of 100,000-fold on average, to the point where error is practically zero. (if you are using the script, you can reduce the error further - we stop the improvement at this point because it is functionally perfect). This means the large over/under-compensation effects can be completely removed from your data. If you want to run 28 colour flow cytometry on a 28 colour machine, you can spend hours-upon-hours compensating your data by hand, or 2 minutes with AutoSpill. AutoSpill is designed to run through the same operations that a skilled flow user does, just faster. But always remember - the compensation can still only be as good as the quality of the single colour controls!
How does AutoSpill work? It is a huge surprise to me, but with the enormous effort over decades to add extra lasers and new flurophores onto machines, the mathematics behind compensation hasn't been updated since 1993, where it was designed for 3 colour flow on computers with 100,000-fold less capacity. For decades we've been building more-and-more expensive machines, and haven't updated the basic maths that the machines run on!
Traditional compensation defines a positive and a negative population, finds the slope and uses that as the spillover matrix. It still works okay in most cases, it is just that the small errors start piling up when you are making 250+ calculations on a high parameter dataset.
AutoSpill is actually fairly simple at heart:
- Draw an automatic live cell gate
- Use linear regression to take into account every cell, not just the two data points of average positive and average negative
- Calculate the error left, using the sum of errors in every compensation pair
- Use the residual error and return to step 2
- Repeat the tweaking of the matrix until error is gone
The underlying mathematics is tougher though (never thought I'd write a paper discussing "the linearity of the quantum mechanical nature of photons"!), because AutoSpill was built for actual flow cytometry users, not as a computational exercise. Carlos built the original pipeline, and then we extensively beta-tested it on 1000+ datasets over 20 months. Something as simple as a live gate becomes complex when you want it to robustly work on any dataset, cells or beads, collected on any machine. I'll spare you the details, but two stage tesselation and a 33% density estimation using a convex hull does the trick, successfully spotting the cells or beads even with heavy debris:
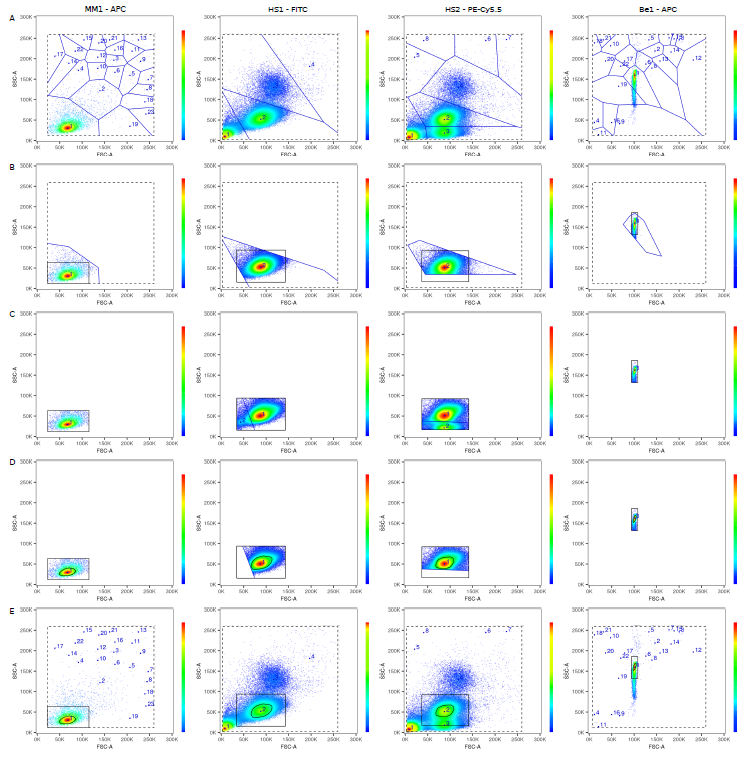
There are many advantages to using linear regression to calculate compensation. Why through out the data from 40,000 cells and instead turn it into two points, the way traditional compensation does? If you use linear regression you can use all of the data, which means AutoSpill works even if you have mostly negative or mostly positive data, just a shoulder of positive events or even a smear. So you can use the real antibodies on real cells to calculate your single colour compensations, rather than using beads or anti-CD4 in every channel.
As an added advantage to this approach, AutoSpill can remove most of the autofluroscence from your flow cytometry sample. For people working on cancer or myeloid cells this can be a complete game-changer. It turns out that while cells have different amounts of autofluorescence, the spectrum of that autofluirescence is fairly constant. You can collect empty data in the worst autofluorescent channel. The single in this channel can be used to calculate the autofluorescent spectrum, which can then be calculated on a per-cell basis and used to compensate it out of every other channel.
Here are two examples:
1. Back when I was a post-doc, there were many published reports of Foxp3 expression in macrophages, epithelium, cancer, etc. All autofluorescent artefacts that wasted years of research.
AutoSpill removes this autofluorescence specifically from the macrophages:
2. Now we work on microglia, and it is still argued in the literature as to whether they express low levels of MHCII at homeostasis. Using AutoSpill to remove the autofluorescent signal it is quite simple - no they don't. 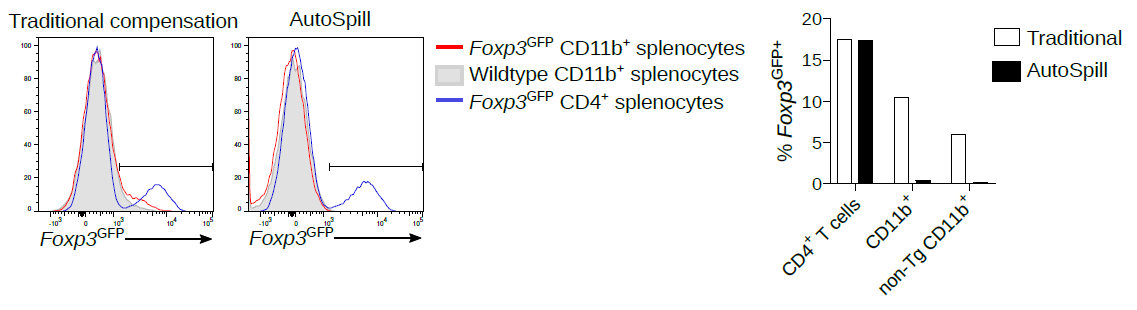
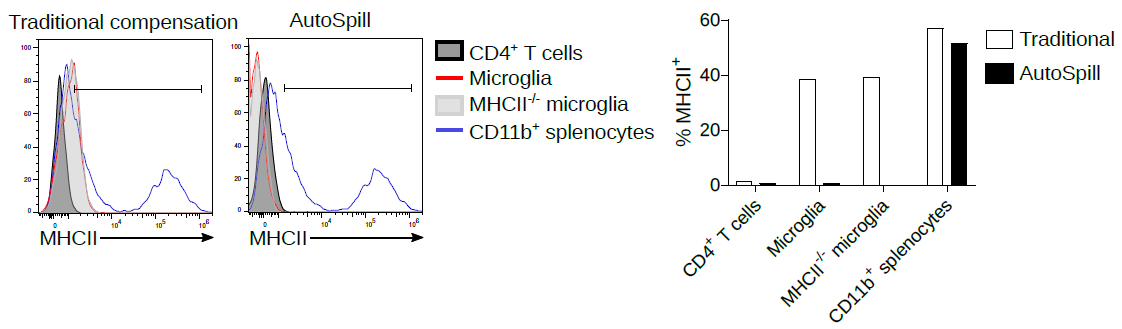
Of course, AutoSpill is a tool to get an optimal solution for good data. It can't turn bad data into good data. You should always work with your Core Facility staff to optimise the machines and run high quality single colour controls.
If you like AutoSpill, and you come from a math, computer science or data science background, why not come and join the lab? We have a position open for a data science post-doc or senior scientist for another two weeks.
 Liston lab,
Liston lab,  data science
data science 




Reader Comments (1)
This post is discussing in an outstanding way. I like your way yo discussed your valid points and facts about this.